Linear Models

import numpy as np
from keras.datasets import mnist
import ssl
ssl._create_default_https_context = ssl._create_unverified_context
from keras.models import Model
from keras.layers import Dense, Input
import matplotlib.pyplot as plt
np.random.seed(9527) # for reproducibility
# Load data
(x_train, _), (x_test, y_test) = mnist.load_data()
image_shape = x_train[0].shape # original image shape
# Create 100 anomalous samples (outliners) in testing data
outlier_index = np.random.randint(0,x_test.shape[0], size=100)
x_test[outlier_index] = x_test[outlier_index] + np.random.normal(0, 40, image_shape)
# Data preprocessing: scaling and reshaping
x_train = x_train.astype('float32') / 255. # minmax_normalized
x_test = x_test.astype('float 32') / 255. # minmax_normalized
x_train = x_train.reshape((x_train.shape[0], -1))
x_test = x_test.reshape((x_test.shape[0], -1))
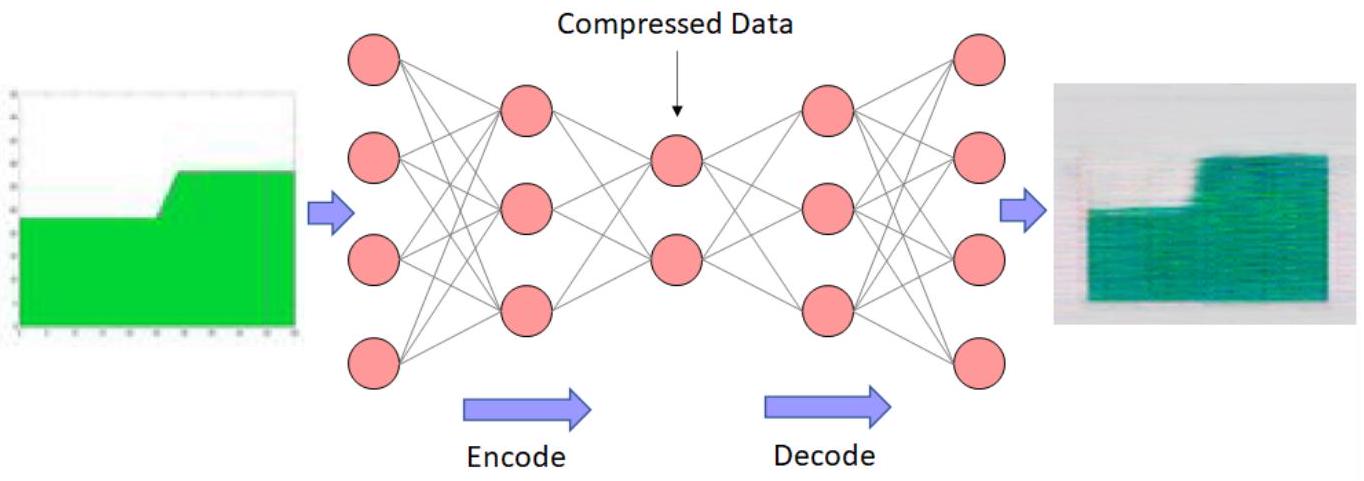
# Built Autoencoder
input_img = Input(shape=(x_train.shape[-1],))
# Encoding
encoded = Dense(24, activation='relu')(input_img)
encoded = Dense(12, activation='relu')(encoded)
encoded = Dense(8, activation='relu') (encoded)
encoder_output = Dense(4)(encoded)
# Decoding
decoded = Dense(8, activation='relu')(encoder_output)
decoded = Dense(12, activation='relu')(decoded)
decoded = Dense(24, activation='relu')(decoded)
decoded = Dense(x_train.shape[-1])(decoded)
# Build autoencoder NN model
autoencoder = Model(inputs=input_img, outputs=decoded)
# Compile autoencoder
autoencoder.compile(loss='mse', optimizer='adam')
# Training
autoencoder.fit(x_train, x_train, epochs=20, batch_size=30, shuffle=True)
# Prediction
autoencoded_imgs = autoencoder.predict(x_test)
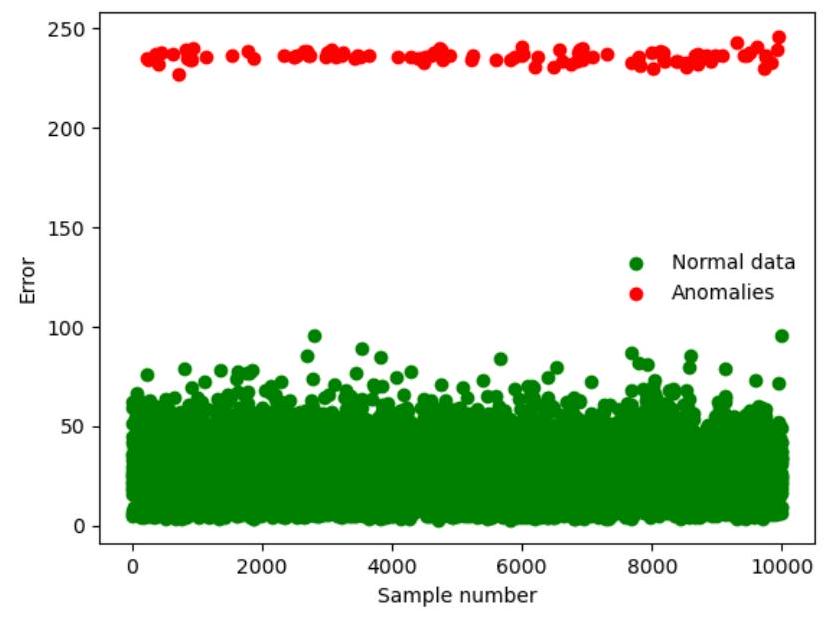
Error = np.sum((x_test-autoencoded_imgs)**2,axis=1) # Sum of squre root errors
Normal_data = np.delete(Error,outlier_index) # Normal data
Outliners = Error[outlier_index] # Anomalies
plt.scatter(np.delete(np.arange(Error.size),outlier_index),Normal_data,c=' g',label='Normal data')
plt.scatter(outlier_index,Error[outlier_index], c='r', label='Anomalies')
plt.legend(loc="middle right",fontsize=10,frameon=False)
plt.xlabel('Sample number')
plt.ylabel('Error')